Artwork © by Greg Findlay
GESTALT: Genome editing of synthetic target arrays for lineage tracing
This is the homepage of GESTALT, the CRISPR/Cas9 lineage tracing approach from the Shendure lab and Schier lab.
Abstract
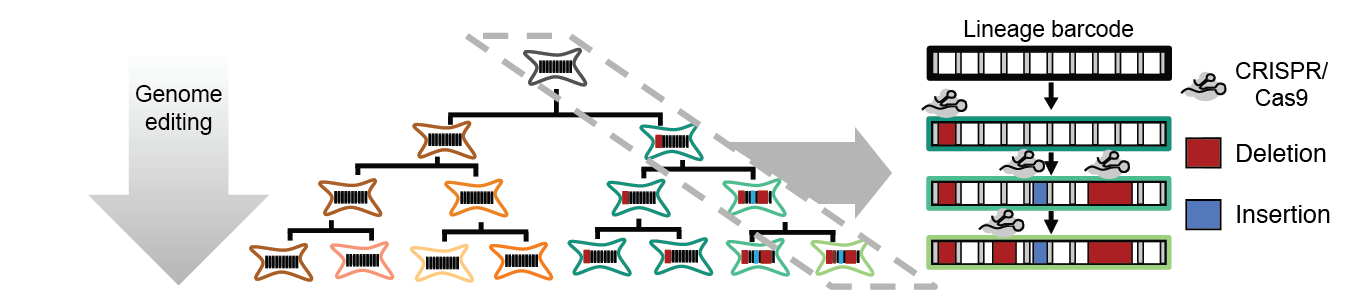
Multicellular systems develop from single cells through a lineage, but current lineage tracing approaches scale poorly to whole organisms. Here we use genome editing to progressively introduce and accumulate diverse mutations in a DNA barcode over multiple rounds of cell division. The barcode, an array of CRISPR/Cas9 target sites, records lineage relationships in the patterns of mutations shared between cells. In cell culture and zebrafish, we show that rates and patterns of editing are tunable, and that thousands of lineage-informative barcode alleles can be generated. By sampling hundreds of thousands of cells from individual zebrafish, we find that most cells in adult zebrafish organs derive from relatively few embryonic progenitors. Genome editing of synthetic target arrays for lineage tracing (GESTALT) will help generate large-scale maps of cell lineage in multicellular systems.
The paper has been published in Science with attached supplemental information. The pre-print is also available on the bioRxiv repository.
Data
The data for the paper is publicly available on both the NCBI's Gene Expression Omnibus website with dataset identifier GSE81713, as well as on the Dryad data repository here. For each sample, both raw reads (in SRA format) as well as statistics (stats) files are included. Let us know if you see anything missing or incomplete and we'll get it fixed.
Stats file description
The stats file contains information about the each individually captured UMI per sample, with the following fields:
- readName - The name of the UMI, including it's UMI sequence and read capture counts
- keep - if the UMI was kept for analysis this field will be PASS, otherwise it will be marked FAIL
- conflict - if the paired-end reads for this UMI couldn't be merged, the two ends are checked for edit consistency. This field can either be marked CONSISTENT or CONFLICTED, in which case the keep field should also be FAIL
- merged - were the paired-end reads for the UMI merged (MERGED) or kept as paired-end (PAIR)?
- target[X] - the called CRISPR editing events over each target cutsite region (where is 1 through the number of targets, between 9 and 12). As described in the methods, CRISPR derived edits that overlap 3 bases up or downstream of the cutsite are counted. Event columns can be NONE when no edit is observed, or contain both the edit length as well as the position (189D+123 means a 189 basepair deletion at position 123 of read when aligned to the reference sequence). Insertions also list the bases in the inserted region like 3I+301+TCA (TCA is inserted here).
Tree data files
In addition for aggregate lineage experiments, like the individual adult fish or the cell culture lineage, trees are available in PHYLIP Mix newick output file (*.newick.txt.gz), as well as our custom JSON file type (*.json.gz) that we adapted for visualization using the Data Driven Documents (D3) library.
Aggregate data
For convenience we've also included aggregated allele files for each of the adult fish, the cell culture data, and the sets of embryos. This files include only UMI tagged amplicons that passed filtering.
- Adult fish ADR1 data with blood alleles in all organs or with blood alleles removed from all organs but blood
- Adult fish ADR2 data with blood alleles in all organs or with blood alleles removed from all organs but blood
- Cell culture alleles
- Embryos carrying the V6 barcode alleles
- Embryos carrying the V7 barcode alleles
Reagents
We've submitted all plasmids (V1-V7) to Addgene, and available on their site at the links below. We've also included links to the full sequence on Benchling.
- Cell culture barcode pLJM1-EGFP-BarcodeV1, full sequence: pLJM1-EGFP-BarcodeV1
- Cell culture barcode pLJM1-EGFP-BarcodeV2, full sequence: pLJM1-EGFP-BarcodeV2
- Cell culture barcode pLJM1-EGFP-BarcodeV3, full sequence: pLJM1-EGFP-BarcodeV3
- Cell culture barcode pLJM1-EGFP-BarcodeV4, full sequence: pLJM1-EGFP-BarcodeV4
- Cell culture barcode pLJM1-EGFP-BarcodeV5, full sequence: pLJM1-EGFP-BarcodeV5
- Adult fish barcode system pTol2-DRv6, full sequence: pTol2-DRv6
- Adult fish barcode system pTol2-DRv7, full sequence: pTol2-DRv7
Code
The code used in the paper is available on the Shendure lab GitHub site here. We've included all the code to process raw reads into event calls, as well as much of the visualization and analysis scripts used in the paper.
Trees
To help users dig into the data, we've created a (very) initial interactive tool for visualizing trees produced in GESTALT. Included are the adult trees highlighted in the paper, the cell culture lineage trees, as well as trees created from individual embryos, many of which didn't make the paper. You can play around with it here.