genomic approaches to developmental biology (selected publications)

Chen, Choi et al. Symbolic recording of signalling and cis-regulatory element activity to DNA. Nature (2024)
PMID:

Hamazaki, Yang et al. Retinoic acid induces human gastruloids with posterior embryo-like structures. Nature Cell Biology (2024)
PMID: 39164488

Qiu, Martin, Welsh et al. A single-cell time-lapse of mouse prenatal development from gastrula to birth. Nature (2024)
PMID: 38355799

Huang, Henck, Qiu et al. Single-cell, whole embryo phenotyping of mammalian developmental disorders. Nature (2023)
(with Spielmann and Cao Labs)
PMID: 37968388

Domcke, Shendure. A reference cell tree will serve science better than a reference cell atlas. Cell (2023)
PMID: 36931241

Chiou, Huang et al. A single-cell multi-omic atlas spanning the adult rhesus macaque brain. Science Advances (2023)
PMID: 37824616

Choi et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature (2022)
PMID: 35794474
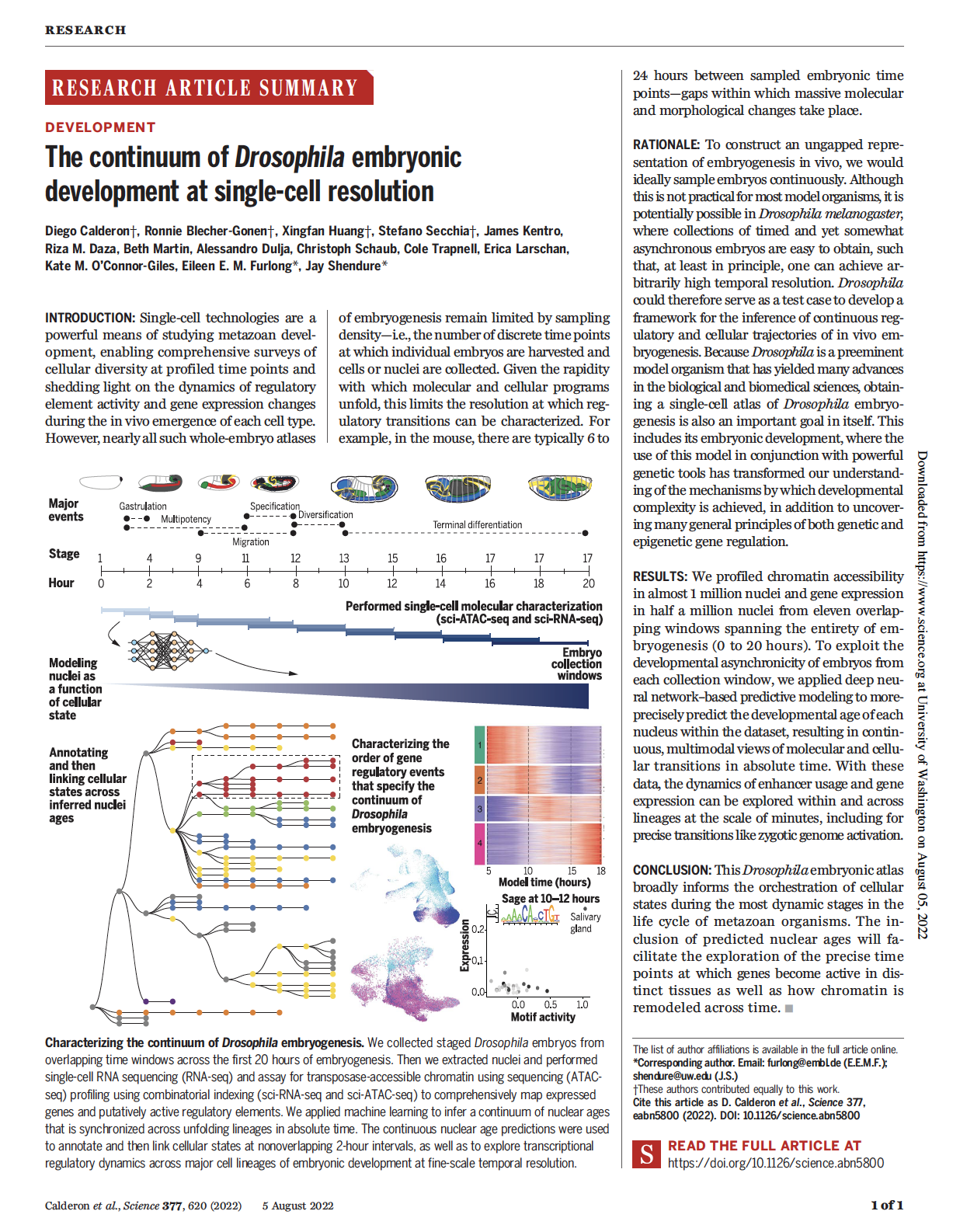
Calderon, Huang et al. The continuum of Drosophila embryonic development at single-cell resolution. Science (2022)
(with Furlong Lab)
PMID: 35926038

Agarwal, Darwin-Lopez et al. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nature Communications (2021)
PMID: 34429411
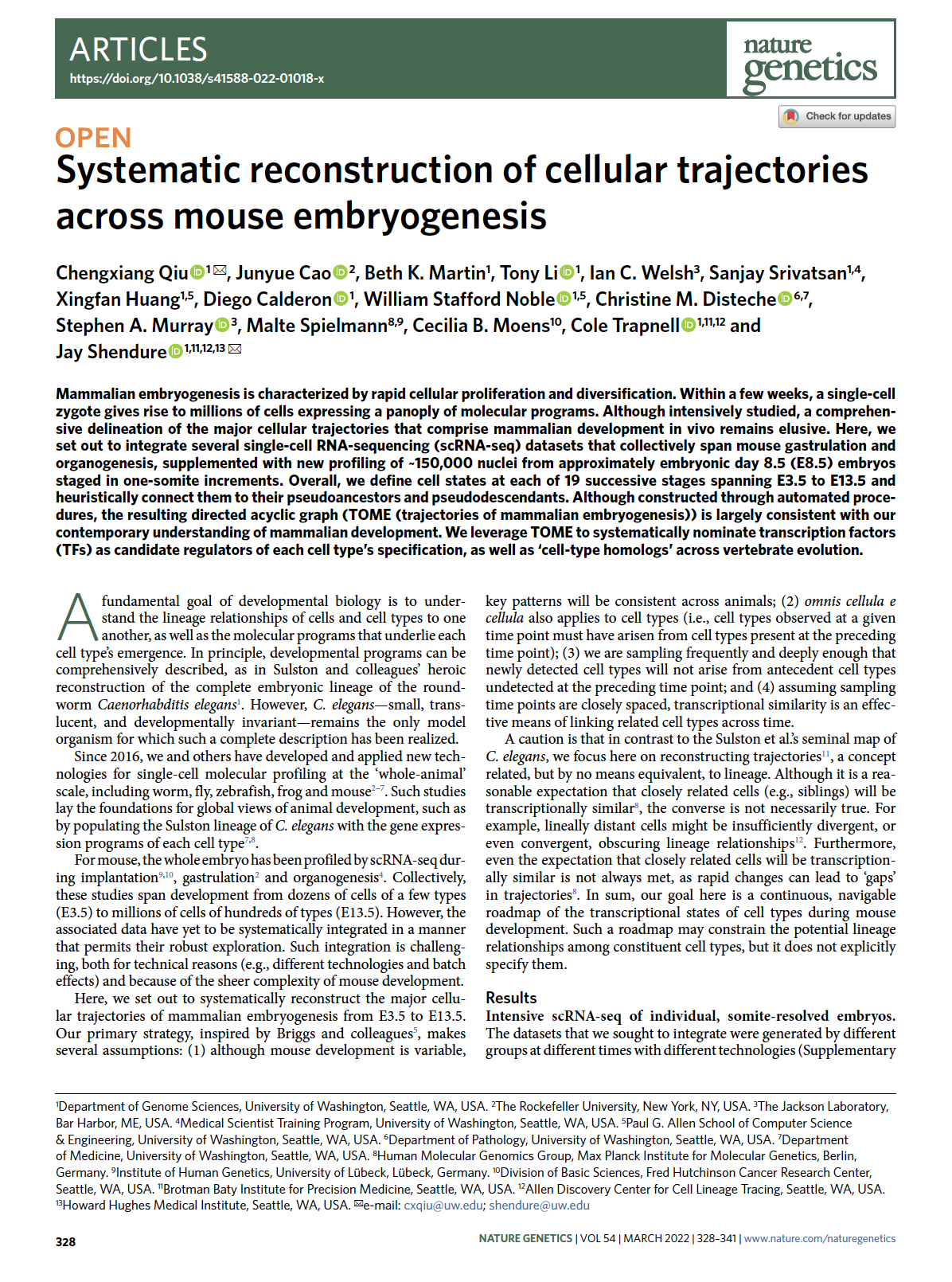
Qiu et al. Systematic reconstruction of cellular trajectories across mouse embryogenesis. Nature Genetics (2022)
PMID: 35288709
.png)
Srivatsan, Regier et al. Embryo-scale, single-cell spatial transcriptomics. Science (2021)
PMID: 34210887
(with Trapnell Lab)
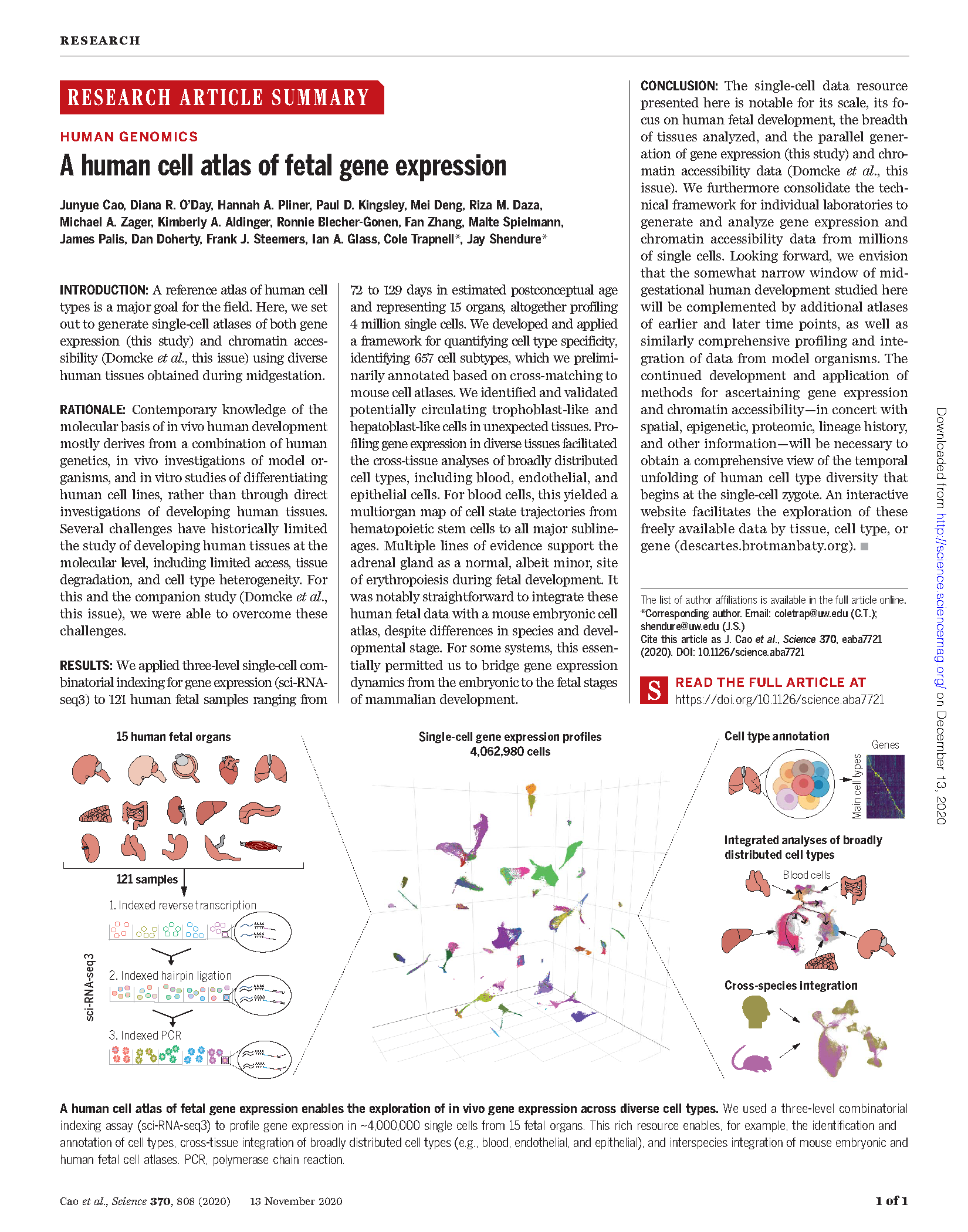
Cao et al. A human cell atlas of fetal gene expression. Science (2020)
PMID: 33184181
(with Trapnell Lab)

Cao et al. Sci-fate characterizes the dynamics of gene expression in single cells. Nature Biotechnology (2020)
PMID: 32284584
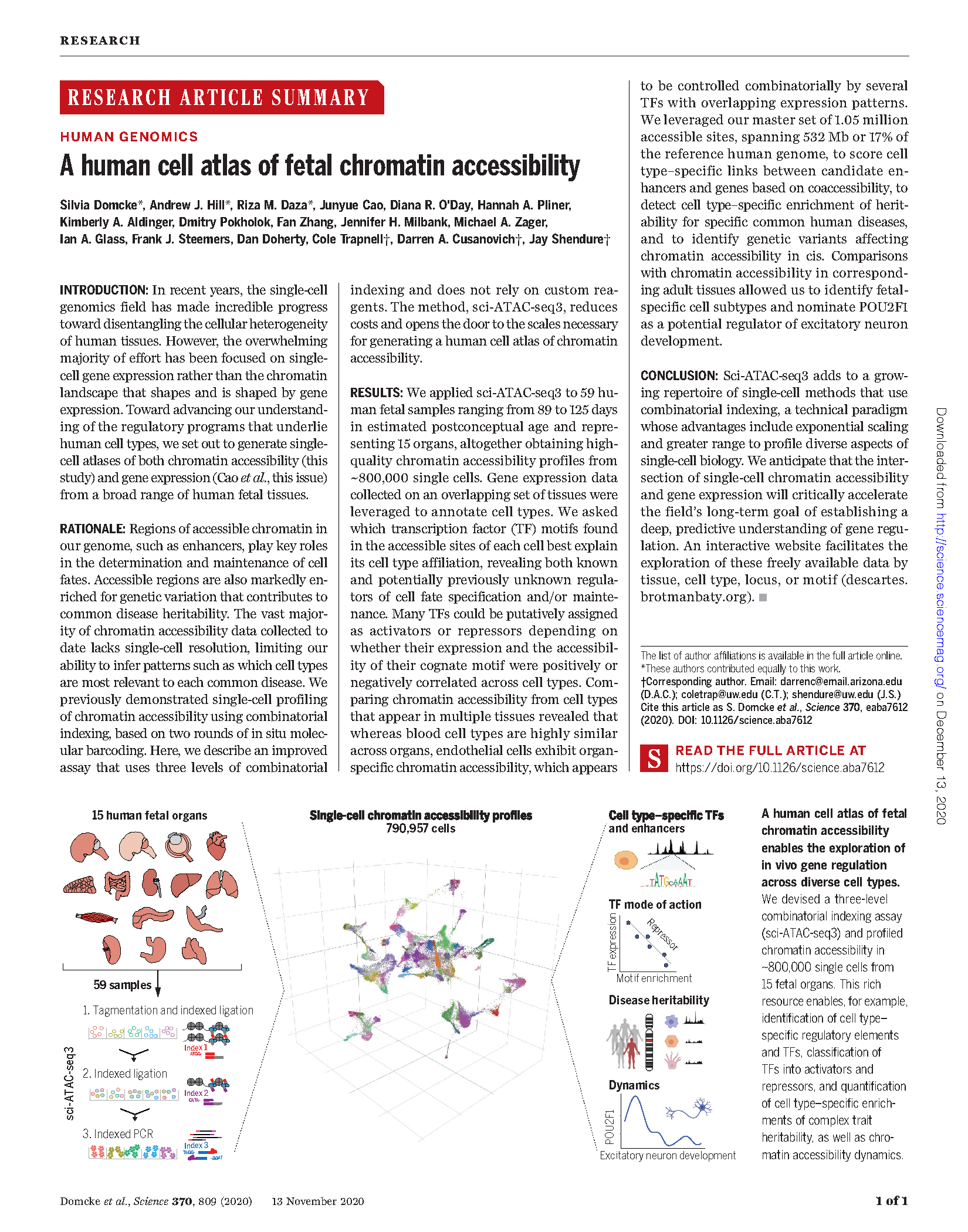
Domcke, Hill, Daza et al. A human cell atlas of fetal chromatin accessibility. Science (2020)
PMID: 33184180
(with Cusanovich and Trapnell Labs)
Yin et al. High-Throughput Single-Cell Sequencing with Linear Amplification. Molecular Cell (2019)
PMID: 31495564
Cao, Spielmann et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature (2019)
PMID: 30787437
(with Trapnell Lab)
Pliner et al. Supervised classification enables rapid annotation of cell atlases. Nature Methods (2019)
PMID: 31501545
(with Trapnell Lab)

Cao et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science (2018)
PMID: 30166440
(with Trapnell Lab)
Pliner et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Molecular Cell (2018)
PMID: 30078726
(with Trapnell Lab)
Cusanovich, Hill et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell (2018)
PMID: 30078704
(with Trapnell Lab)
Cusanovich, Reddington, Garfield et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature (2018)
PMID: 29539636
(with Furlong Lab)
McKenna, Findlay, Gagnon et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science (2016)
PMID: 27229144
(with Schier Lab)

Cao, Packer et al. Comprehensive single cell transcriptional profiling of a multicelluar organism by combinatorial indexing. Science (2017)
PMID: 28818938
(with Trapnell and Waterston Labs)
