developing new molecular methods (selected publications)
Choi et al. A molecular proximity sensor based on an engineered, dual-component guide RNA. eLife (2025)
PMID: 39937081

McDiarmid, Taylor et al. A parts list of promoters and gRNA scaffolds for mammalian genome engineering and molecular recording. Nature Biotechnology (2025)
PMID: 41219485

Chen, Choi et al. Symbolic recording of signalling and cis-regulatory element activity to DNA. Nature (2024)
PMID: 39020177

Li et al. Chromatin context-dependent regulation and epigenetic manipulation of prime editing. Cell (2024)
PMID: 37090511

Liao, Choi, Shendure. Molecular recording using DNA Typewriter. Nature Protocols (2024)
PMID: 38844553

Martin et al. Optimized single-nucleus transcriptional profiling by combinatorial indexing. Nature Protocols (2022)
PMID: 36261634
Choi, Chen et al. Precise genomic deletions using paired prime editing. Nature Biotechnology (2022)
PMID: 34650269

Choi et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature (2022)
PMID: 35794474
.png)
Srivatsan, Regier et al. Embryo-scale, single-cell spatial transcriptomics. Science (2021)
PMID: 34210887
(with Trapnell and Stevens Labs)
Domcke, Hill, Cusanovich, Daza et al. A human cell atlas of fetal chromatin accessibility. Science (2020)
PMID: 33184180
(with Cusanovich and Trapnell Labs)
.png)
Simeonov et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell (2021)
PMID: 34115987
(with Lengner and McKenna Labs)

Cao et al. Sci-fate characterizes the dynamics of gene expression in single cells. Nature Biotechnology (2020)
PMID: 32284584
Yin et al. High-Throughput Single-Cell Sequencing with Linear Amplification. Molecular Cell (2019)
PMID: 31495564

Srivatsan et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science (2020)
PMID: 31806696
(with Trapnell Lab)
Alexander et al. Concurrent genome and epigenome editing by CRISPR-mediated sequence replacement. BMC Biology (2019)
PMID: 31739790
Kircher, Xiong, Martin, Schubach et al. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nature Communications (2019)
PMID: 31395865
(with Ahituv and Kircher Labs)
Pliner et al. Supervised classification enables rapid annotation of cell atlases. Nature Methods (2019)
PMID: 31501545
(with Trapnell Lab)

Chen et al. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Research (2019)
PMID: 31165867

Ramani et al. High Sensitivity Profiling of Chromatin Structure by MNase-SSP. Cell Reports (2019)
PMID: 30811994

Kim et al. A combination of transcription factors mediates inducible interchromosomal contacts. eLife (2019)
PMID: 31081754
Cao, Spielmann et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature (2019)
PMID: 30787437

Cao et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science (2018)
PMID: 30166440
(with Trapnell Lab)

Gasperini et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell (2019)
PMID: 30612741
Matreyek, Starita et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nature Genetics (2018)
PMID: 29785012
(with Fowler Lab)

Cao, Packer et al. Comprehensive single cell transcriptional profiling of a multicelluar organism by combinatorial indexing. Science (2017)
PMID: 28818938
(with Trapnell and Waterston Labs)
Hill, McFaline-Figueroa et al. On the design of CRISPR-based single-cell molecular screens. Nature Methods (2018)
PMID: 29457792
(with Trapnell Lab)

Ramani, Cusanovich et al. Mapping 3D genome architecture through in situ DNase Hi-C. Nature Protocols (2016)
PMID: 27685100

Gasperini, Findlay et al. CRISPR/Cas9-Mediated Scanning for Regulatory Elements Required for HPRT1 Expression via Thousands of Large, Programmed Genomic Deletions. AJHG (2017)
PMID: 28712454
McKenna, Findlay, Gagnon et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science (2016)
PMID: 27229144
(with Schier Lab)
Ramani et al. High-throughput determination of RNA structure by proximity ligation. Nature Biotechnology (2015)
PMID: 26237516

Klein et al. Multiplex pairwise assembly of array-derived DNA oligonucleotides. Nucleic Acids Research (2015)
PMID: 26553805

Cusanovich et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (2015)
PMID: 25953818
(with Trapnell Lab)

Snyder et al. Haplotype-resolved genome sequencing: experimental methods and applications. Nature Reviews Genetics (2015)
PMID: 25948246

Kitzman, Starita et al. Massively parallel single-amino-acid mutagenesis. Nature Methods (2015)
PMID: 25559584
(with Fields Lab)

Boyle et al. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics (2014)
PMID: 24867941

Hiatt et al. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Research (2013)
PMID: 23382536
Shendure, Lieberman-Aiden. The expanding scope of DNA sequencing. Nature Biotechnology (2012)
PMID: 23138308

Adey, Shendure. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Research (2012)
PMID: 22466172

Adey, Morrison, Asan, Xun et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biology (2010)
PMID: 21143862
(with Zhang Lab)
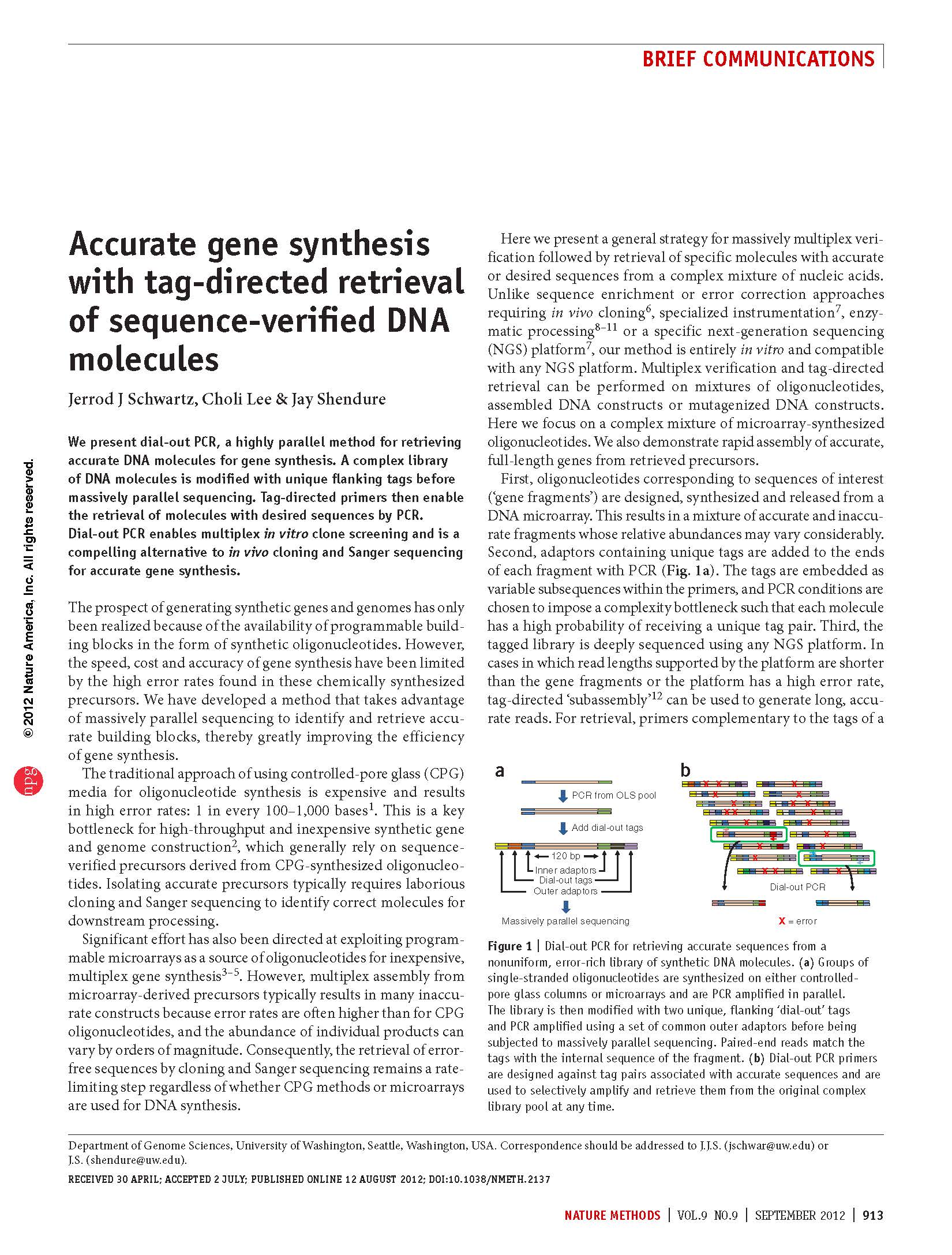
Schwartz et al. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nature Methods (2012)
PMID: 22886093
Hiatt, Patwardhan et al. Parallel, tag-directed assembly of locally derived short sequence reads. Nature Methods (2010)
PMID: 20081835
Porreca, Zhang et al. Multiplex Amplification of Large Sets of Human Exons. Nature Methods (2007)
PMID: 17934468
(in Church Lab)
Turner et al. Massively parallel exon capture and library-free resequencing across 16 genomes. Nature Methods (2009)
PMID: 19349981

