featured research articles (work led or co-led by our lab)

McDiarmid, Taylor et al. A parts list of promoters and gRNA scaffolds for mammalian genome engineering and molecular recording. Nature Biotechnology (2025)
PMID: 41219485
Choi et al. A molecular proximity sensor based on an engineered, dual-component guide RNA. eLife (2025)
PMID: 39937081
Pinglay et al. Multiplex generation and single-cell analysis of structural variants in mammalian genomess. Science (2025)
PMID: 38405830
Suiter et al. Combinatorial mapping of E3 ubiquitin ligases to their target substrates. Molecular Cell (2025)
PMID: 39919746

Agarwal, Inoue et al. Massively parallel characterization of transcriptional regulatory elements. Nature (2025)
PMID: 39814889

Hamazaki, Yang et al. Retinoic acid induces human gastruloids with posterior embryo-like structures. Nature Cell Biology (2024)
PMID: 39164488
Chardon, McDiarmid et al. Multiplex, single-cell CRISPRa screening for cell type specific regulatory elements. Nature Communications (2024)
PMID: 39294132

Chen, Choi et al. Symbolic recording of signalling and cis-regulatory element activity to DNA. Nature (2024)
PMID: 39020177

Li et al. Chromatin context-dependent regulation and epigenetic manipulation of prime editing. Cell (2024)
PMID: 37090511

Lalanne, Regalado et al. Multiplex profiling of developmental cis-regulatory elements with quantitative single-cell expression reporters. Nature Methods (2024)
PMID: 38724692

Qiu, Martin, Welsh et al. A single-cell time-lapse of mouse prenatal development from gastrula to birth. Nature (2024)
PMID: 38355799

Huang, Henck, Qiu et al. Single-cell, whole embryo phenotyping of mammalian developmental disorders. Nature (2023)
(with Spielmann and Cao Labs)
PMID: 37968388
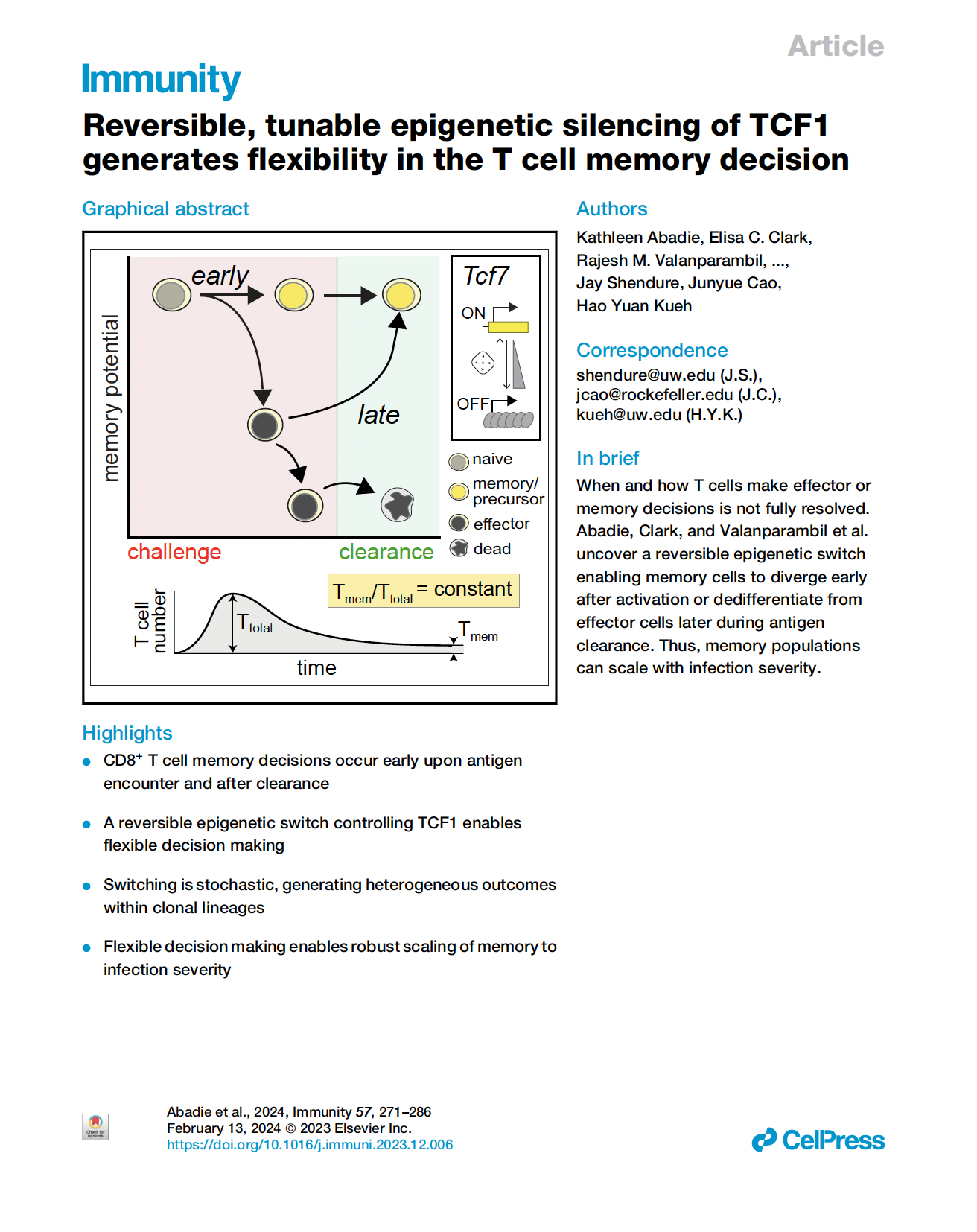
Abadie, Clark, Valanparambil et al. Reversible, tunable epigenetic silencing of TCF-1 generates flexibility in the T cell memory decision. Cell (2024)
(with Kueh Lab)
PMID: 38301652

Chiou, Huang et al. A single-cell multi-omic atlas spanning the adult rhesus macaque brain. Science Advances (2023)
(with Snyder-Mackler and Platt Labs)
PMID: 37824616
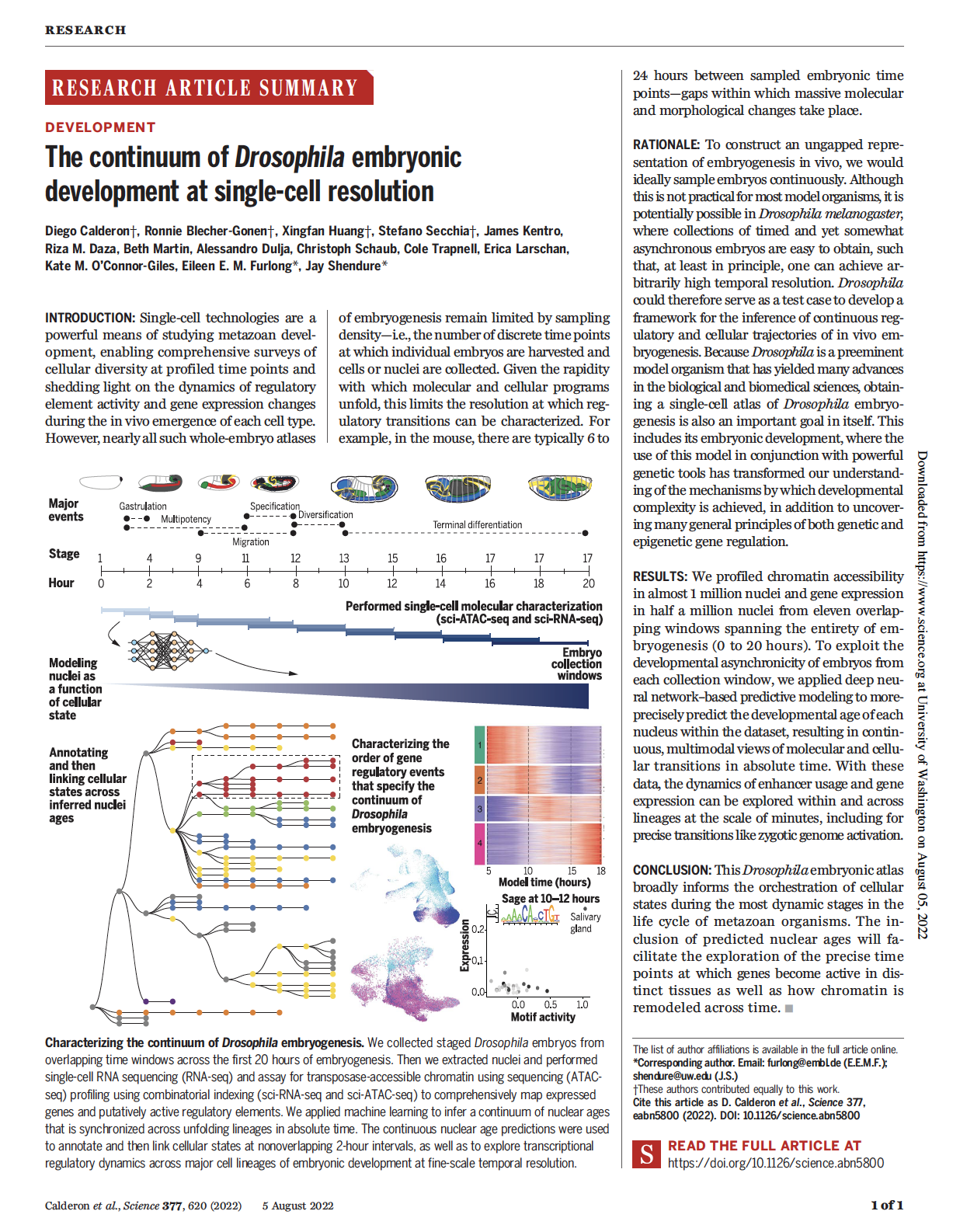
Calderon, Huang et al. The continuum of Drosophila embryonic development at single-cell resolution. Science (2022)
(with Furlong Lab)
PMID: 35926038

Martin et al. Optimized single-nucleus transcriptional profiling by combinatorial indexing. Nature Protocols (2022)
PMID: 36261634

Choi et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature (2022)
PMID: 35794474
Qiu et al. Systematic reconstruction of cellular trajectories across mouse embryogenesis. Nature Genetics (2022)
PMID: 35288709
Choi, Chen et al. Precise genomic deletions using paired prime editing. Nature Biotechnology (2022)
PMID: 34650269

Agarwal, Darwin-Lopez et al. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nature Communications (2021)
PMID: 34429411
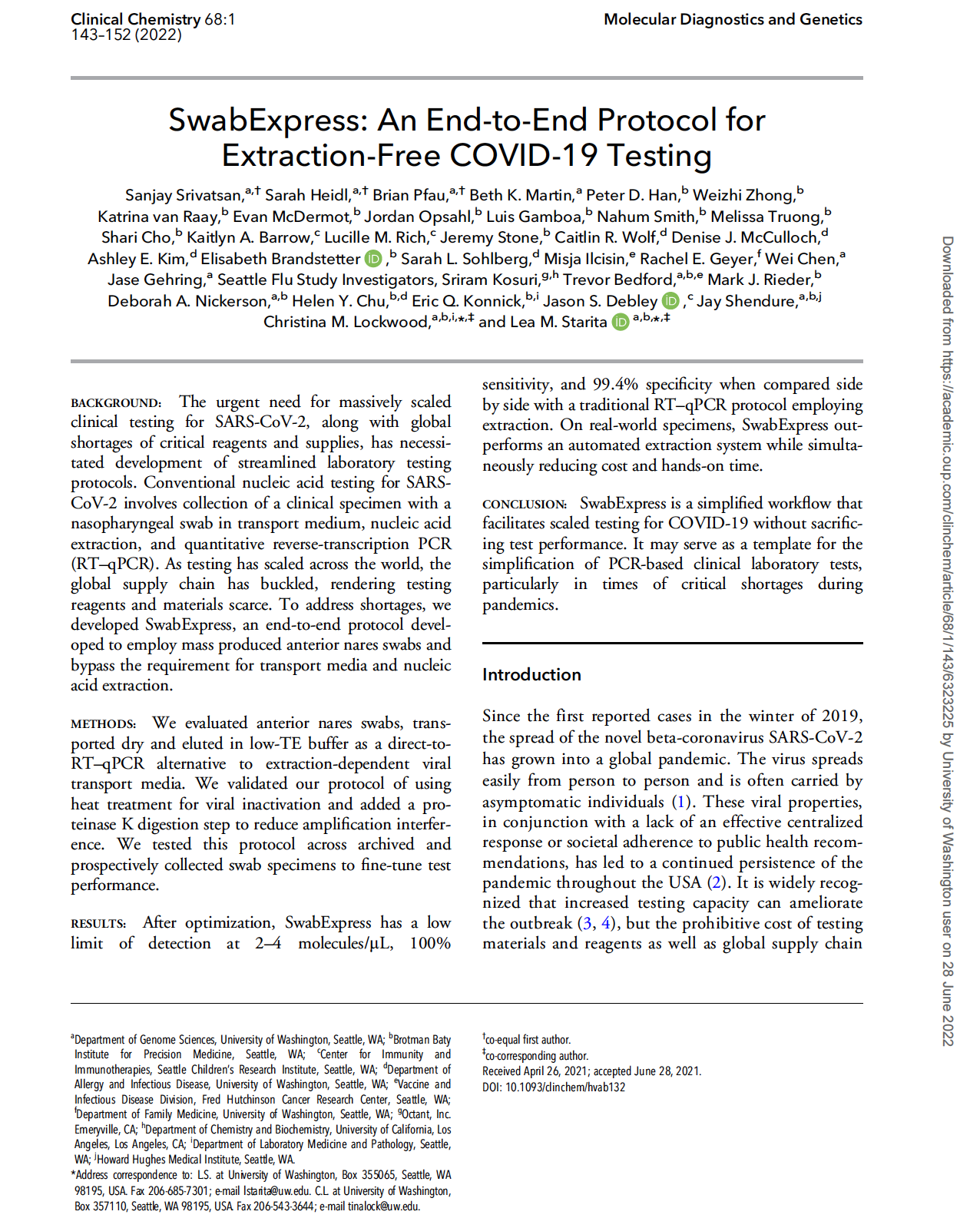
Srivatsan et al. SwabExpress: An end-to-end protocol for extraction-free COVID-19 testing. Clinical Chemistry (2021)
PMID: 32511368
(with Seattle Flu Study)
.png)
Srivatsan, Regier et al. Embryo-scale, single-cell spatial transcriptomics. Science (2021)
PMID: 34210887
(with Trapnell and Stevens Labs)
.png)
Simeonov et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell (2021)
PMID: 34115987
(with Lengner and McKenna Labs)
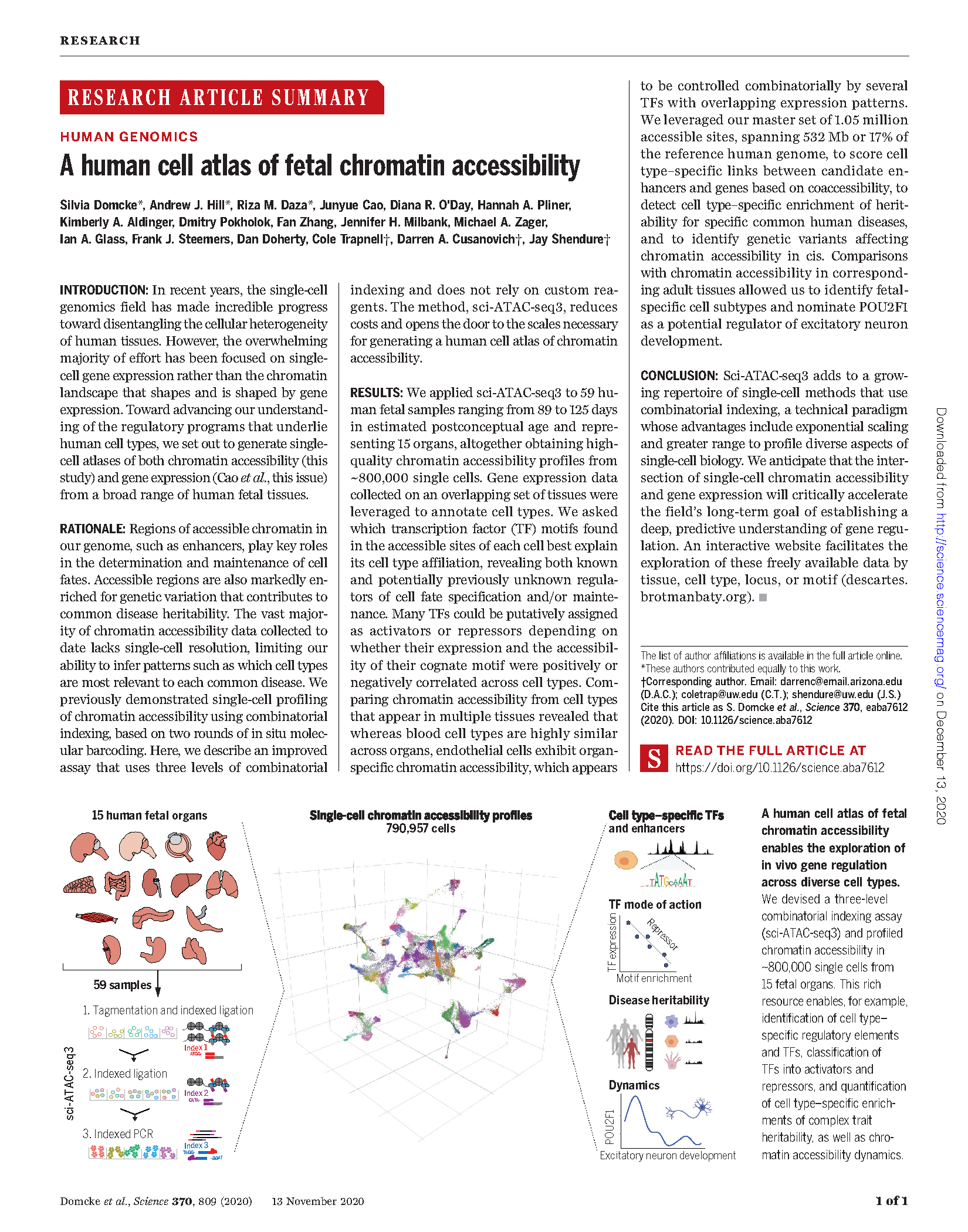
Domcke, Hill, Daza et al. A human cell atlas of fetal chromatin accessibility. Science (2020)
PMID: 33184180
(with Cusanovich and Trapnell Labs)
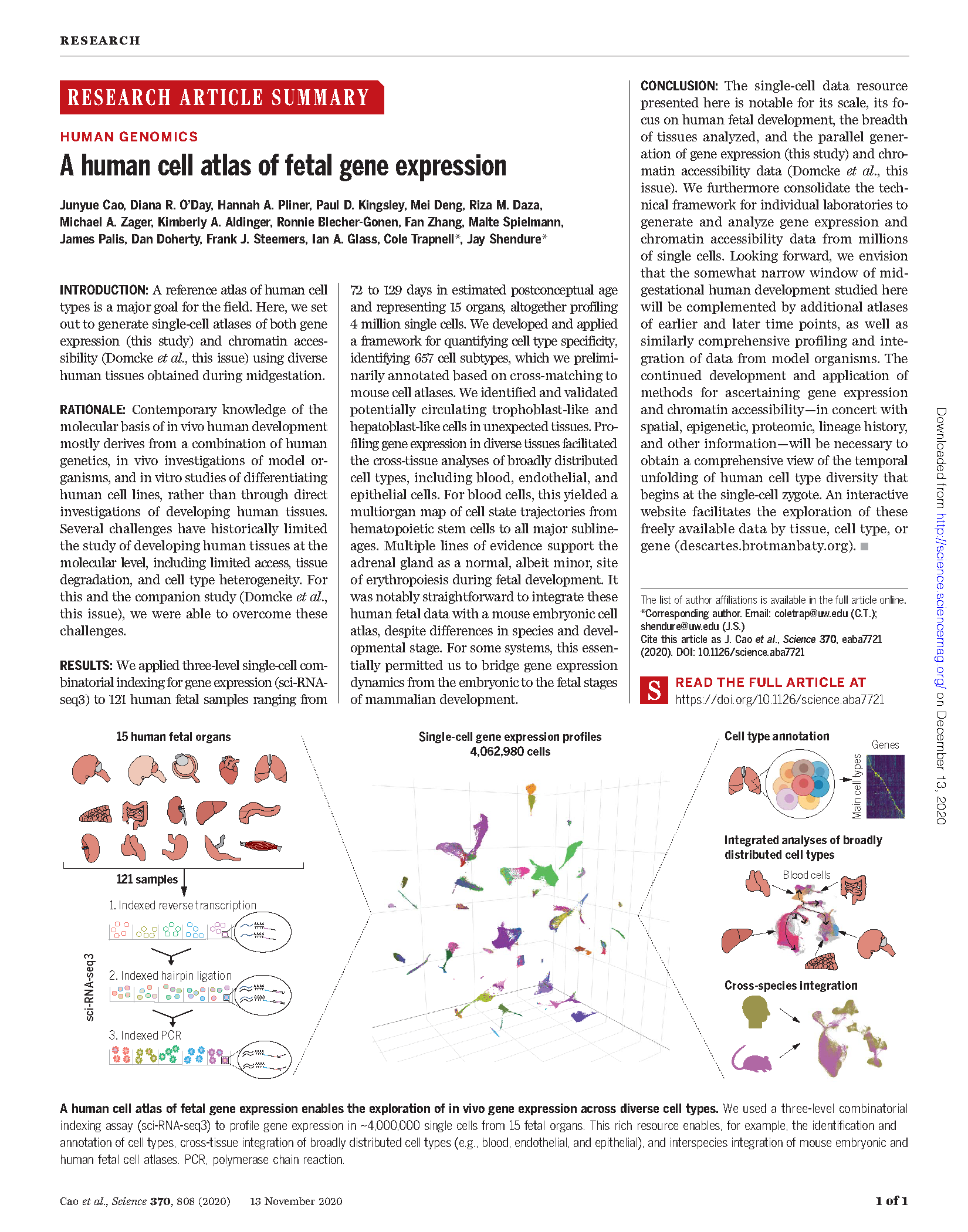
Cao et al. A human cell atlas of fetal gene expression. Science (2020)
PMID: 33184181
(with Trapnell Lab)
Klein, Agarwal, Inoue, Keith et al. A systematic evaluation of the design and context dependencies of massively parallel reporter assays. Nature Methods (2020)
PMID: 33046894
(with Ahituv Lab)
Chu et al. Early Detection of Covid-19 through a Citywide Pandemic Surveillance Platform. New England Journal of Medicine (2020)
PMID: 32356944
(with Seattle Flu Study)

Bedford, Greninger,Roychoudhury, Starita, Famulare et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science (2020)
PMID: 32511596
(with Seattle Flu Study)

Cao et al. Sci-fate characterizes the dynamics of gene expression in single cells. Nature Biotechnology (2020)
PMID: 32284584
(with Trapnell Lab)

Srivatsan, McFaline-Figueroa, Ramani et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science (2020)
PMID: 31806696
(with Trapnell Lab)
Agarwal, Shendure. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks. Cell Reports (2020)
PMID: 32433972
Yin et al. High-Throughput Single-Cell Sequencing with Linear Amplification. Molecular Cell (2019)
PMID: 31495564
Pliner et al. Supervised classification enables rapid annotation of cell atlases. Nature Methods (2019)
PMID: 31501545
(with Trapnell Lab)
Alexander et al. Concurrent genome and epigenome editing by CRISPR-mediated sequence replacement. BMC Biology (2019)
PMID: 31739790
Kircher, Xiong, Martin, Schubach et al. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nature Communications (2019)
PMID: 31395865
(with Ahituv and Kircher Labs)

Klein, Keith et al. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. Nature Communications (2019)
PMID: 31164647

Chen, McKenna, Schrieber et al. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Research (2019)
PMID: 31165867

Kim et al. A combination of transcription factors mediates inducible interchromosomal contacts. eLife (2019)
PMID: 31081754
Cao, Spielmann et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature (2019)
PMID: 30787437
(with Trapnell Lab)

Ramani et al. High Sensitivity Profiling of Chromatin Structure by MNase-SSP. Cell (2019)
PMID: 30811994

Gasperini et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell (2019)
PMID: 30612741

Starita et al. A Multiplex Homology-Directed DNA Repair Assay Reveals the Impact of More Than 1,000 BRCA1 Missense Substitution Variants on Protein Function. AJHG (2018)
PMID: 30219179
(with Parvin Lab)

Rentzsch et al. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research (2019)
PMID: 30371827
(with Kircher Lab)
Findlay et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature (2018)
PMID: 30209399
(with Starita Lab)
Pliner et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Molecular Cell (2018)
PMID: 30078726
(with Trapnell Lab)

Cao et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science (2018)
PMID: 30166440
(with Trapnell Lab)
Cusanovich, Hill et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell (2018)
PMID: 30078704
(with Trapnell Lab)
Cusanovich, Reddington, Garfield et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature (2018)
PMID: 29539636
(with Furlong Lab)
McKenna & Shendure. FlashFry: a fast and flexible tool for large-scale CRISPR target design. BMC Biology (2018)
PMID: 29976198
Klein et al. Functional characterization of enhancer evolution in the primate lineage. Genome Biology (2018)
PMID: 30045748

Cao, Packer et al. Comprehensive single cell transcriptional profiling of a multicelluar organism by combinatorial indexing. Science (2017)
PMID: 28818938
(with Trapnell and Waterston Labs)
Hill, McFaline-Figueroa et al. On the design of CRISPR-based single-cell molecular screens. Nature Methods (2018)
PMID: 29457792
(with Trapnell Lab)
Matreyek, Starita et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nature Genetics (2018)
PMID: 29785012
(with Fowler Lab)

Gasperini, Findlay et al. CRISPR/Cas9-Mediated Scanning for Regulatory Elements Required for HPRT1 Expression via Thousands of Large, Programmed Genomic Deletions. AJHG (2017)
PMID: 28712454

Kim et al. The dynamic three-dimensional organization of the diploid yeast genome. eLIFE (2017)
PMID: 28537556
(with Dunham Lab)

Neveling, Mensenkamp et al. BRCA Testing by Single-Molecule Molecular Inversion Probes. Clinical Chemistry (2017)
PMID: 27974384
(with Nelen and Hoischen Labs)
Hause et al. Classification and characterization of microsatellite instability across 18 cancer types. Nature Medicine (2016)
PMID: 27694933
(with Salipante Lab)

Inoue, Kircher et al. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Research (2017)
PMID: 27831498
(with Ahituv Lab)
Snyder, Kircher et al. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell (2016)
PMID: 26771485

Ramani, Cusanovich et al. Mapping 3D genome architecture through in situ DNase Hi-C. Nature Protocols (2016)
PMID: 27685100

Kumar, Coleman et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nature Medicine (2016)
PMID: 26928463
(with Nelson Lab)
Ramani et al. High-throughput determination of RNA structure by proximity ligation. Nature Biotechnology (2015)
PMID: 26237516
McKenna, Findlay, Gagnon et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science (2016)
PMID: 27229144
(with Schier Lab)

Underhill et al. Fragment Length of Circulating Tumor DNA. PLoS Genetics (2016)
PMID: 27428049
(with Underhill Lab)

Starita et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics (2015)
PMID: 25823446
(with Fields Lab)

Fairfield et al. Exome sequencing reveals pathogenic mutations in 91 strains of mice with Mendelian disorders. Genome Research (2015)
PMID: 25917818
(with Reinholdt Lab)

Klein et al. Multiplex pairwise assembly of array-derived DNA oligonucleotides. Nucleic Acids Research (2015)
PMID: 26553805

Kitzman, Starita et al. Massively parallel single-amino-acid mutagenesis. Nature Methods (2015)
PMID: 25559584
(with Fields Lab)

Roach et al. A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota. PLoS Genetics (2015)
PMID: 26230489
(with Salipante Lab)

Cusanovich et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (2015)
PMID: 25953818
(with Trapnell Lab)

O'Roak, Stessman et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nature Communications (2014)
PMID: 25418537
(with Eichler Lab)
Kumar, Ryan et al. Whole genome prediction for preimplantation genetic diagnosis. Genome Medicine (2015)
PMID: 26019723

Snyder, Simmons et al. Copy-Number Variation and False Positive Prenatal Aneuploidy Screening Results. New England Journal of Medicine (2015)
PMID: 25830323
(with Gammill Lab)

Salipante, Roach et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Research (2014)
PMID: 25373147
Kumar et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biology (2014)
PMID: 25608559
(with Rostomily Lab)

Findlay, Boyle et al. Saturation editing of genomic regions by multiplex homology-directed repair. Nature (2014)
PMID: 25141179

Burton, Liachko et al. Species-Level Deconvolution of Metagenome Assemblies with Hi-C–Based Contact Probability Maps. G3 (2014)
PMID: 24855317
(with Dunham Lab)
Adey et al. In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Research (2014)
PMID: 25327137
Iossifov, O'Roak, Sanders, Ronemus et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature (2014)
PMID: 25363768
(with Wigler, State and Eichler Labs)

Boyle et al. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics (2014)
PMID: 24867941

Schwartz, Roach et al. Primate evolution of the recombination regulator PRDM9. Nature Communications (2014)
PMID: 25001002
Kircher, Witten et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics (2014)
PMID: 24487276
(with Witten and Cooper Labs)

Hiatt et al. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Research (2013)
PMID: 23382536
Burton et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature Biotechnology (2013)
PMID: 24185095

Adey, Burton, Kitzman et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature (2013)
PMID: 23925245

O'Roak et al. Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders. Science (2012)
PMID: 23160955
(with Eichler Lab)
Smith, Taher L et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nature Genetics (2013)
(with Ahituv and Ovcharenko Labs)
PMID: 23892608
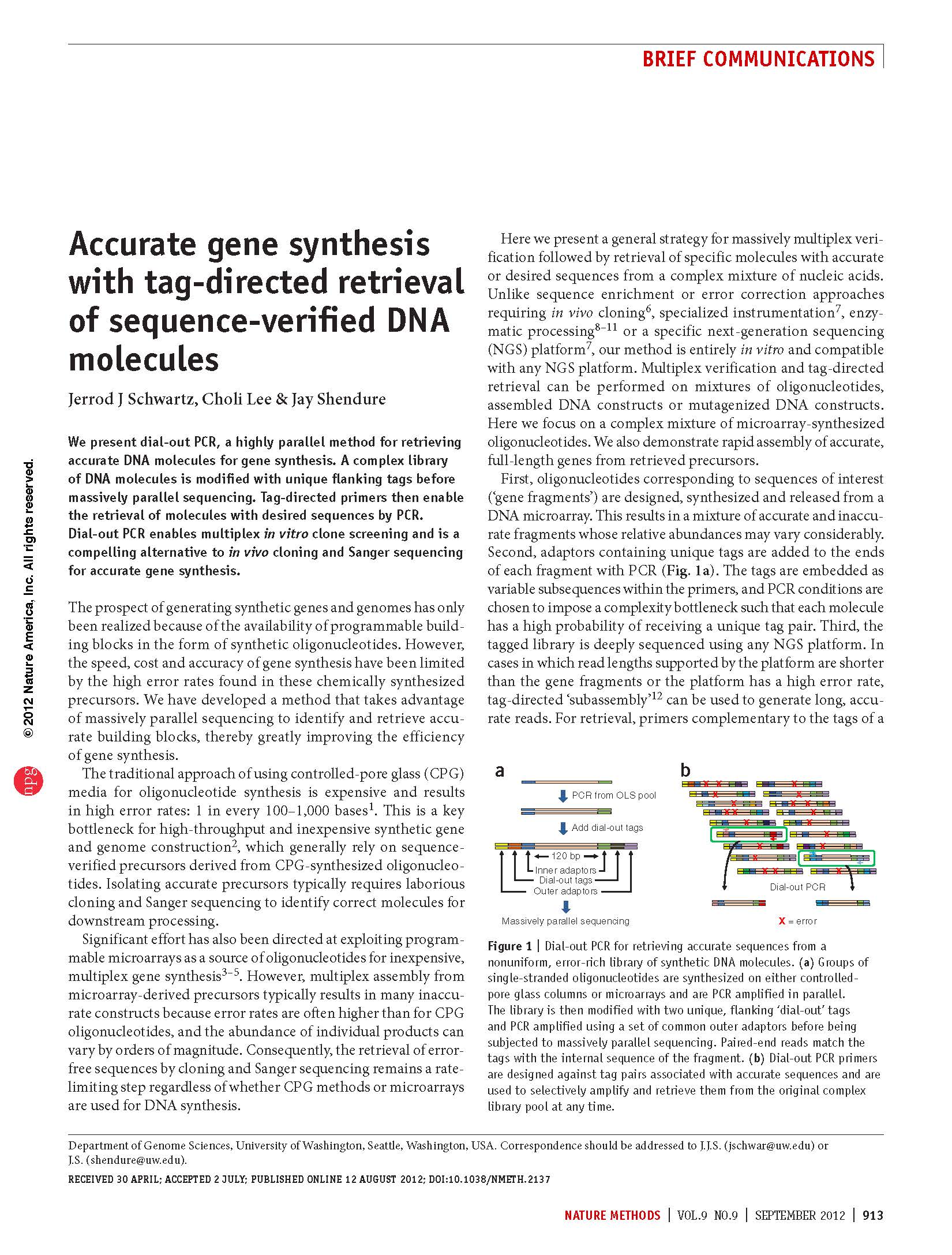
Schwartz et al. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nature Methods (2012)
PMID: 22886093
Patwardhan, Hiatt et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature Biotechnology (2012)
PMID: 22371081
(with Pennacchio and Ahituv Labs)

Schwartz et al. Capturing native long-range contiguity by in situ library construction and optical sequencing. PNAS (2012)
PMID: 23112150

Kitzman et al. Noninvasive whole-genome sequencing of a human fetus. Science Translational Medicine (2012)
PMID: 22674554

Adey, Shendure. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Research (2012)
PMID: 22466172

O'Roak et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature (2012)
PMID: 22495309
(with Eichler Lab)

Kumar et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. PNAS (2011)
PMID: 21949389
(with Nelson Lab)
O'Roak et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics (2011)
PMID: 21572417
(with Eichler Lab)

George et al. Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Research (2011)
PMID: 21795384
(with Thomas Lab)

Kitzman et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nature Biotechnology (2011)
PMID: 21170042
Hiatt, Patwardhan et al. Parallel, tag-directed assembly of locally derived short sequence reads. Nature Methods (2010)
PMID: 20081835
Ng, Bigham et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature Genetics (2010)
PMID: 20711175
(with Bamshad Lab)

Adey, Morrison, Asan, Xun et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biology (2010)
PMID: 21143862
(with Zhang Lab)
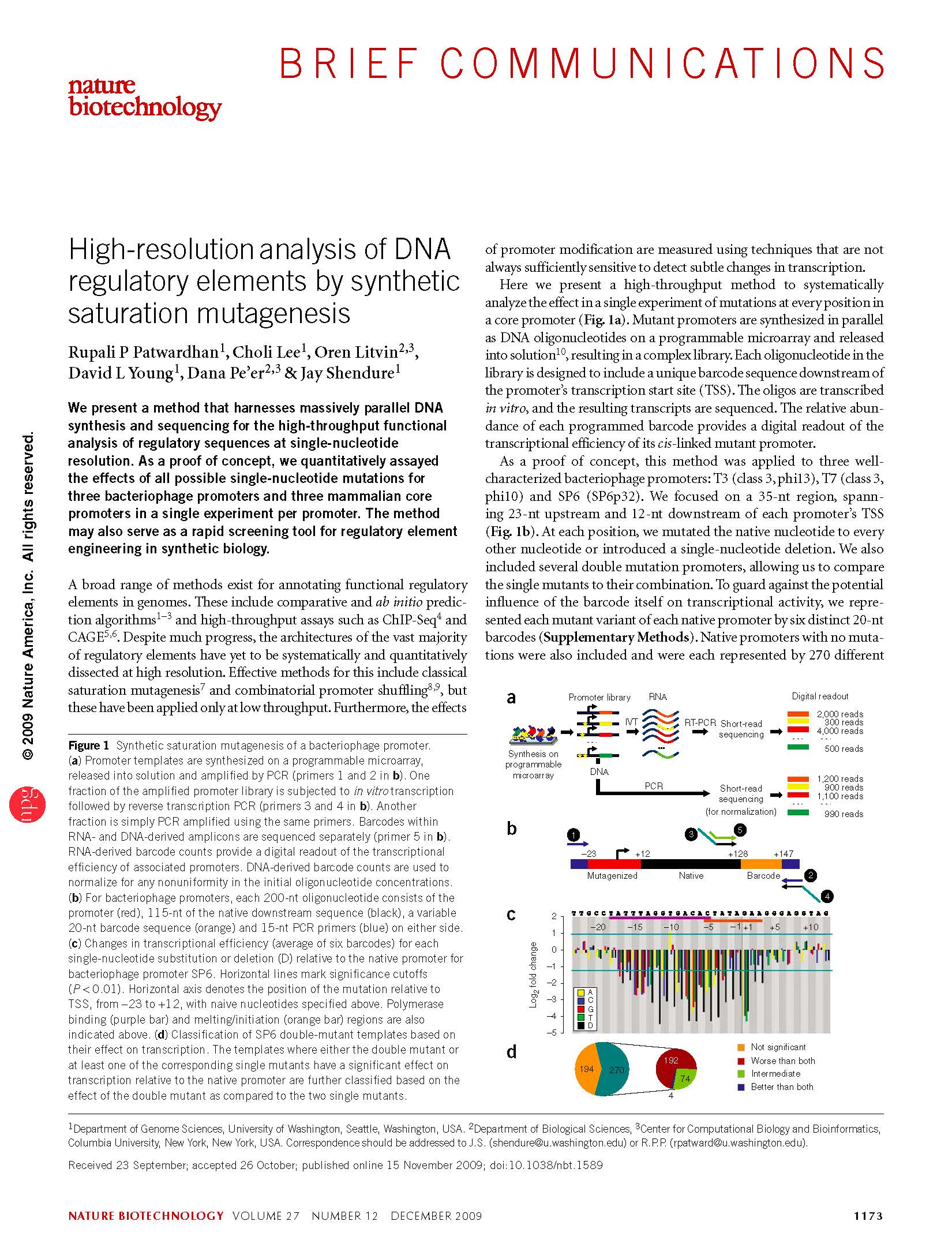
Patwardhan et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nature Biotechnology (2009)
PMID: 19915551
Ng, Buckingham et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genetics (2010)
PMID: 19915526
(with Bamshad Lab)

Ng et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature (2009)
PMID: 19684571

Vasta V et al. Next generation sequence analysis for mitochondrial disorders. Genome Medicine (2009)
PMID: 19852779
(with Hahn Lab)
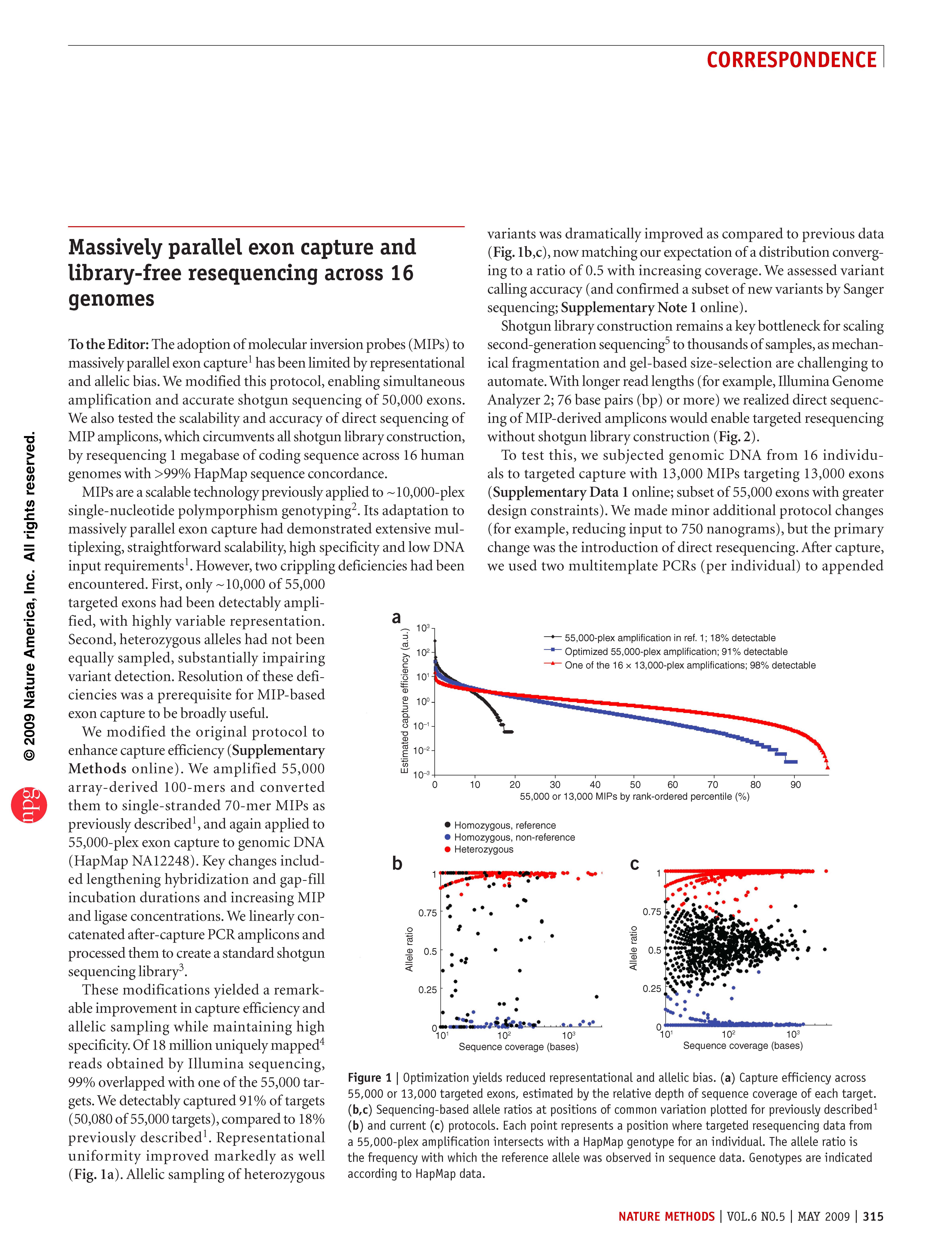
Turner et al. Massively parallel exon capture and library-free resequencing across 16 genomes. Nature Methods (2009)
PMID: 19349981

