massively parallel functional genomics (selected publications)
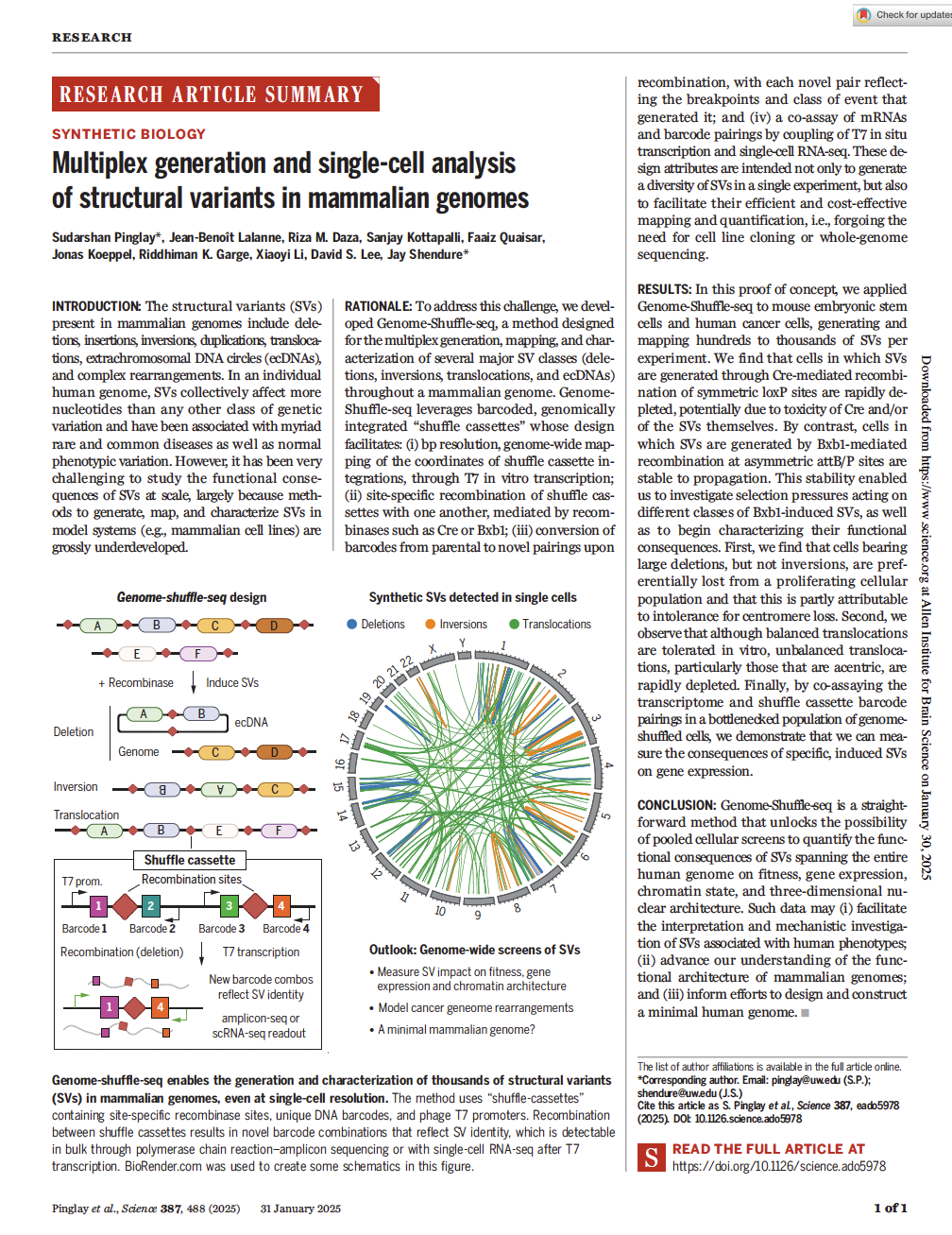
Pinglay et al. Multiplex generation and single-cell analysis of structural variants in mammalian genomess. Science (2025)
PMID: 38405830
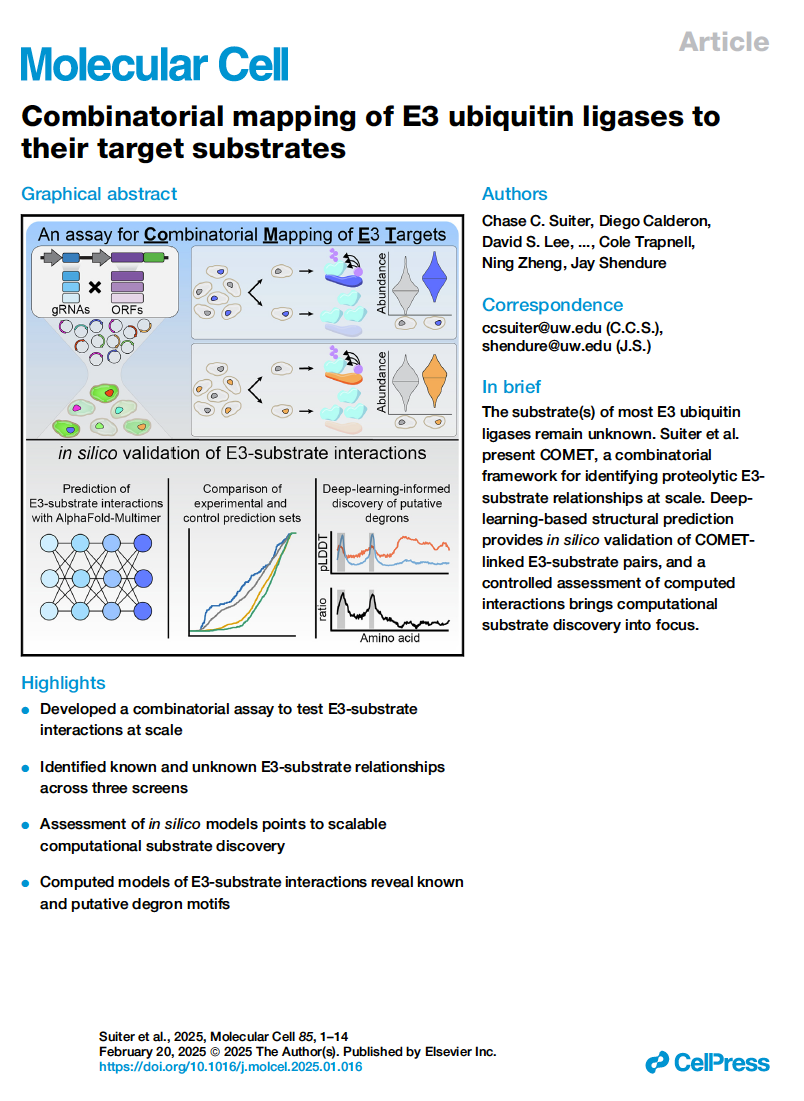
Suiter et al. Combinatorial mapping of E3 ubiquitin ligases to their target substrates. Molecular Cell (2025)
PMID: 39919746

Agarwal, Inoue et al. Massively parallel characterization of transcriptional regulatory elements. Nature (2025)
PMID: 39814889

Lalanne, Regalado et al. Multiplex profiling of developmental cis-regulatory elements with quantitative single-cell expression reporters. Nature Methods (2024)
PMID: 38724692
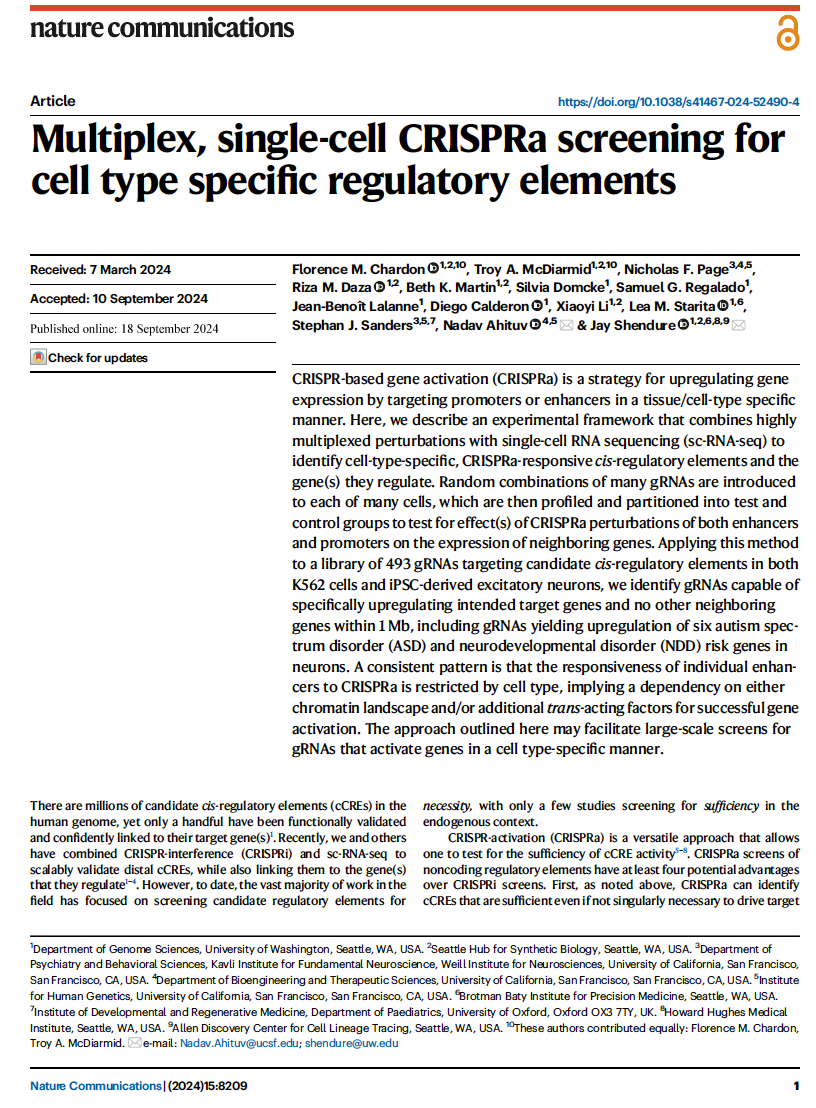
Chardon, McDiarmid et al. Multiplex, single-cell CRISPRa screening for cell type specific regulatory elements. Nature Communications (2024)
PMID: 39294132
Klein, Agarwal, Inoue, Keith et al. A systematic evaluation of the design and context dependencies of massively parallel reporter assays. Nature Methods (2020)
PMID: 33046894

Gasperini, Tome, Shendure. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nature Reviews Genetics (2020)
PMID: 31988385

Gordon, Inoue, Martin, Schubach et al. lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nature Protocols (2020)
PMID: 32641802
(with Ahituv and Kircher Labs)

Srivatsan, McFaline-Figueroa, Ramani et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science (2020)
PMID: 31806696
(with Trapnell Lab)

Chen, McKenna, Schreiber et al. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Research (2019)
PMID: 31165867
Kircher, Xiong, Martin, Schubach et al. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nature Communications (2019)
PMID: 31395865
(with Ahituv and Kircher Labs)

Klein, Keith et al. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. Nature Communications (2019)
PMID: 31164647

Gasperini et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell (2019)
PMID: 30612741

Kim, Dunham, Shendure. A combination of transcription factors mediates inducible interchromosomal contacts. eLife (2019)
PMID: 31081754
Findlay et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature (2018)
PMID: 30209399
(with Starita Lab)

Starita et al. A Multiplex Homology-Directed DNA Repair Assay Reveals the Impact of More Than 1,000 BRCA1 Missense Substitution Variants on Protein Function. AJHG (2018)
PMID: 30219179
(with Parvin Lab)
Klein et al. Functional characterization of enhancer evolution in the primate lineage. Genome Biology (2018)
PMID: 30045748
Hill, McFaline-Figueroa et al. On the design of CRISPR-based single-cell molecular screens. Nature Methods (2018)
PMID: 29457792
(with Trapnell Lab)

Gasperini, Findlay et al. CRISPR/Cas9-Mediated Scanning for Regulatory Elements Required for HPRT1 Expression via Thousands of Large, Programmed Genomic Deletions. AJHG (2017)
PMID: 28712454
Matreyek, Starita et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nature Genetics (2018)
PMID: 29785012
(with Fowler Lab)

Inoue, Kircher et al. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Research (2017)
PMID: 27831498
(with Ahituv Lab)
Gasperini, Starita, Shendure. The power of multiplexed functional analysis of genetic variants. Nature Protocols (2016)
PMID: 27583640

Starita, Shendure, Fields et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics (2015)
PMID: 25823446
(with Fields Lab)

Findlay, Boyle et al. Saturation editing of genomic regions by multiplex homology-directed repair. Nature (2014)
PMID: 25141179

Kitzman, Starita et al. Massively parallel single-amino-acid mutagenesis. Nature Methods (2015)
PMID: 25559584
(with Fields Lab)
Smith, Taher L et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nature Genetics (2013)
PMID: 23892608
(with Ahituv, Ovcharenko Labs)
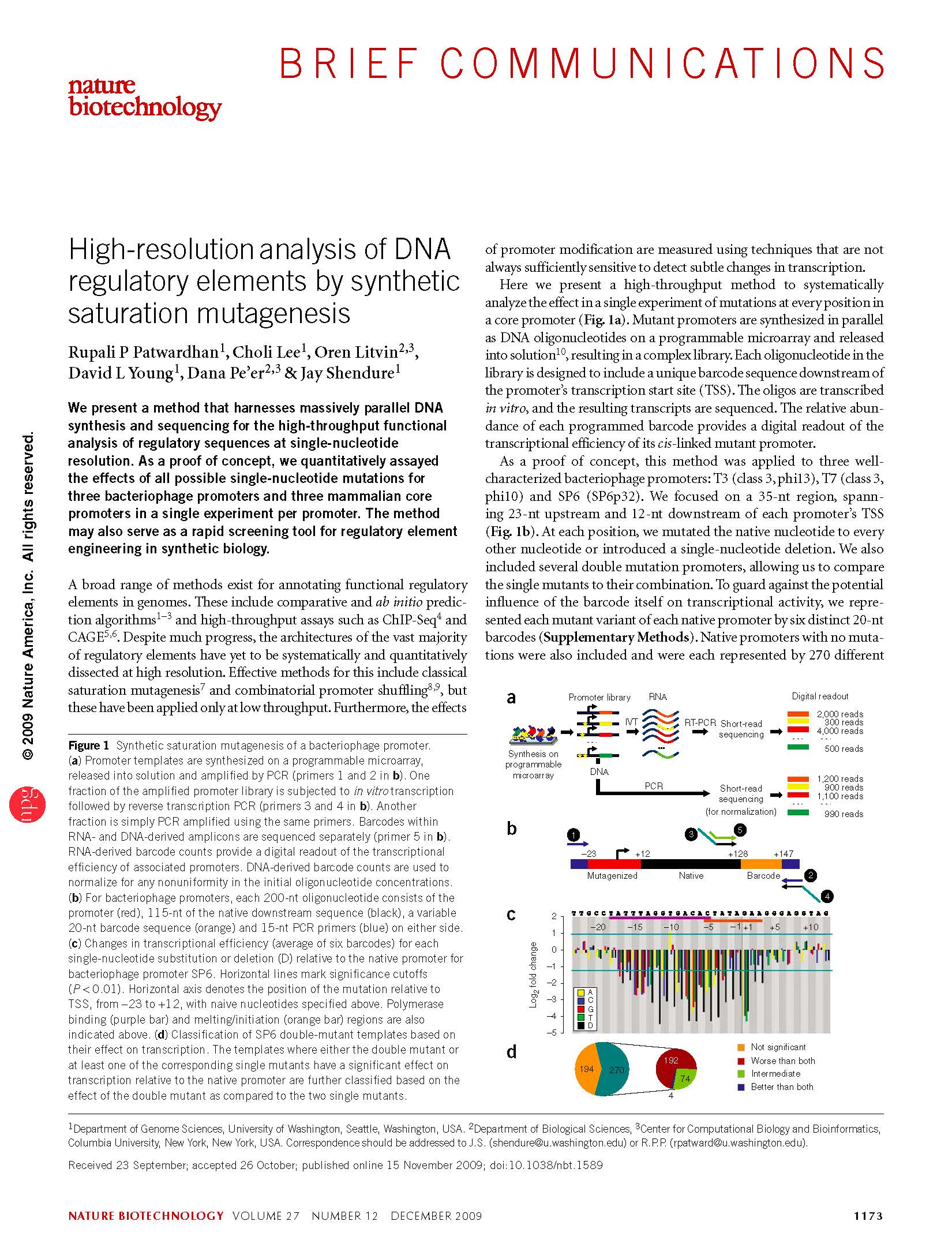
Patwardhan et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nature Biotechnology (2009)
PMID: 19915551
Patwardhan, Hiatt et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature Biotechnology (2012)
PMID: 22371081
(with Pennacchio, Ahituv Labs)